Intro
They say most of the time spent, when working in Data Science, is spent on preparing the data for the  analysis. This has definitely been my experience so far.
analysis. This has definitely been my experience so far.
That one time, not so long ago, a Wednesday night, I was working on a report – actually I made it a Dashboard (yes, Shiny). The report was due the next morning (last minute task finding its way to the top of the list… Anyway). Because of the urgency, at some point I had tested ingesting some of the data, and I was already advancing with the rest of the script, when I realised I was going to need to read in MANY files (about one year’s worth of hourly data tables).
From past experiences I imagined it would take quite a few minutes more for the execution, and it was getting late… Until I thought I would test reading in all the data before going any further. After a “few minutes”, I concluded that reading in the data would take no less than 3 hours… I really didn’t have that kind of time.
So I needed a solution, and I needed it fast. Of course I wasn’t the first one to face such a problem, and StackOverflow, once again, came to the rescue.
Here I explain a demo (you can find it here on my github account) I just created, to present a similar hypothetical situation, and the solution I used then (I now only rarely use any other way of reading in many CSV files).
The right tool for the task
Well, as it turns out, maybe R is not the BEST option here.
For instance, it should be fairly easy in bash to put back together many CSV files into one bigger CSV, and then read that. But even then, reading in a VERY big CSV might prove a bit slow with read.csv().
As it turns out, at least whenever I have used and compared the two, data.table’s fread() has proven (much) faster than read.csv(). So that was one thing right there to improve execution times.
But then, there was the rbind() part.
Reading in many files, and doing one rbind() EVERY TIME I read in a file, in a for(){} loop, that was sub-optimal to say the least. Something about loading into memory more data with each iteration… If only there was a way to put together MANY data.frames in one go… Someone was nice enough to explain just that in a post in r-bloggers already, and as they mention: “The classic rbind in a for loop is massively influenced by the number of sub-dataframes!”
Which was my experience. To demo that, I modified the script (the one I published here for this precise blog post) to plot the execution time between each two iterations of the for loop.
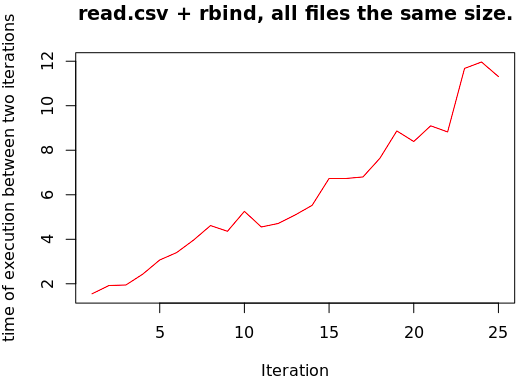
Now you can tell (obviously) the line isn’t straight and horizontal, but rather increasing (i.e. exponentially slower, the more files I read in). However, the file size in each iteration is essentially constant. This is NOT GOOD if you are going to read in MANY files into one data.frame…
What about the style
As per the “style of the code”, well… I wasn’t too fond of the “apply()” family of functions for a long time (I kind of defaulted to for(){} loops for years, which is what I knew from programming in C mainly, back at the university) and that alone was a handicap.
Notwithstanding the execution time, I used a for loop, where I could use an apply function which usually makes the code cleaner.
In summary
Reading many CSV files in a folder, before:
tempdf <- read.csv(file = files[1], header = FALSE, sep = "\t")
for(i in files[2:num_files]) {
tempdf <- rbind(tempdf,
read.csv(i, header = FALSE, sep = "\t"))
}
After:
tempdf2 <- rbind.fill(lapply(files[1:num_files], fread, header = FALSE, sep = "\t"))
And to clarify: On my machine (and my Docker Container), after a few tests, to read in 10 files of 100.000 lines each, the second option was approximately 10 times faster each time.
References
https://www.r-bloggers.com/the-rbinding-race-for-vs-do-call-vs-rbind-fill/
https://stackoverflow.com/questions/31316343/reading-multiple-csv-files-faster-into-data-table-r
https://github.com/kaizen-R/R/blob/master/Sample/demos/comparing_reading_speeds_v001.R